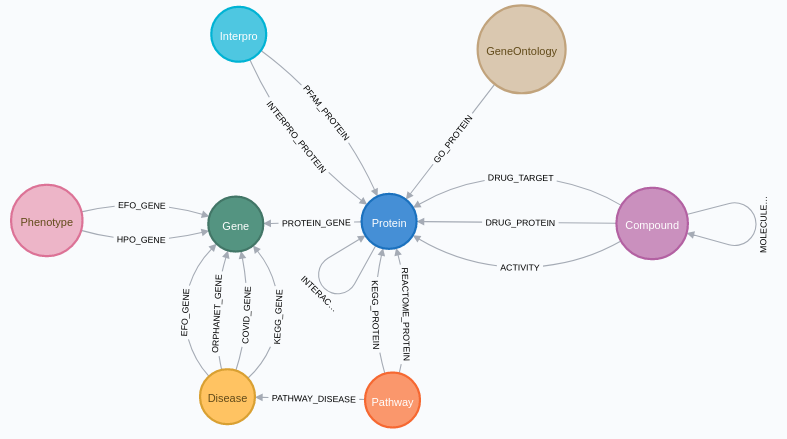
SmartBioGraph Database
SmartBioGraph Database (SmartBioGraphDB) is a graph database mainly based on the data retrieved from CROssBAR database, which is a document-oriented database. Currently, SmartBioGraphDB has over 2.5 million nodes with 16 labels and over 11 million relationships with 17 types. Our node types that come from CROssBAR are proteins (UniProt), genes (UniProt and HPO gene symbols), compounds (CHEMBL and DrugBank), pathways (Reactome), diseases (EFO), phenotypes (HPO), Gene Ontology and Interpro nodes. In addition to nodes coming from CROssBAR database, we also provide disease and pathway nodes which come from KEGG. There are also cross-references for nodes to other databases such as PubChem, OMIM and PubMed.
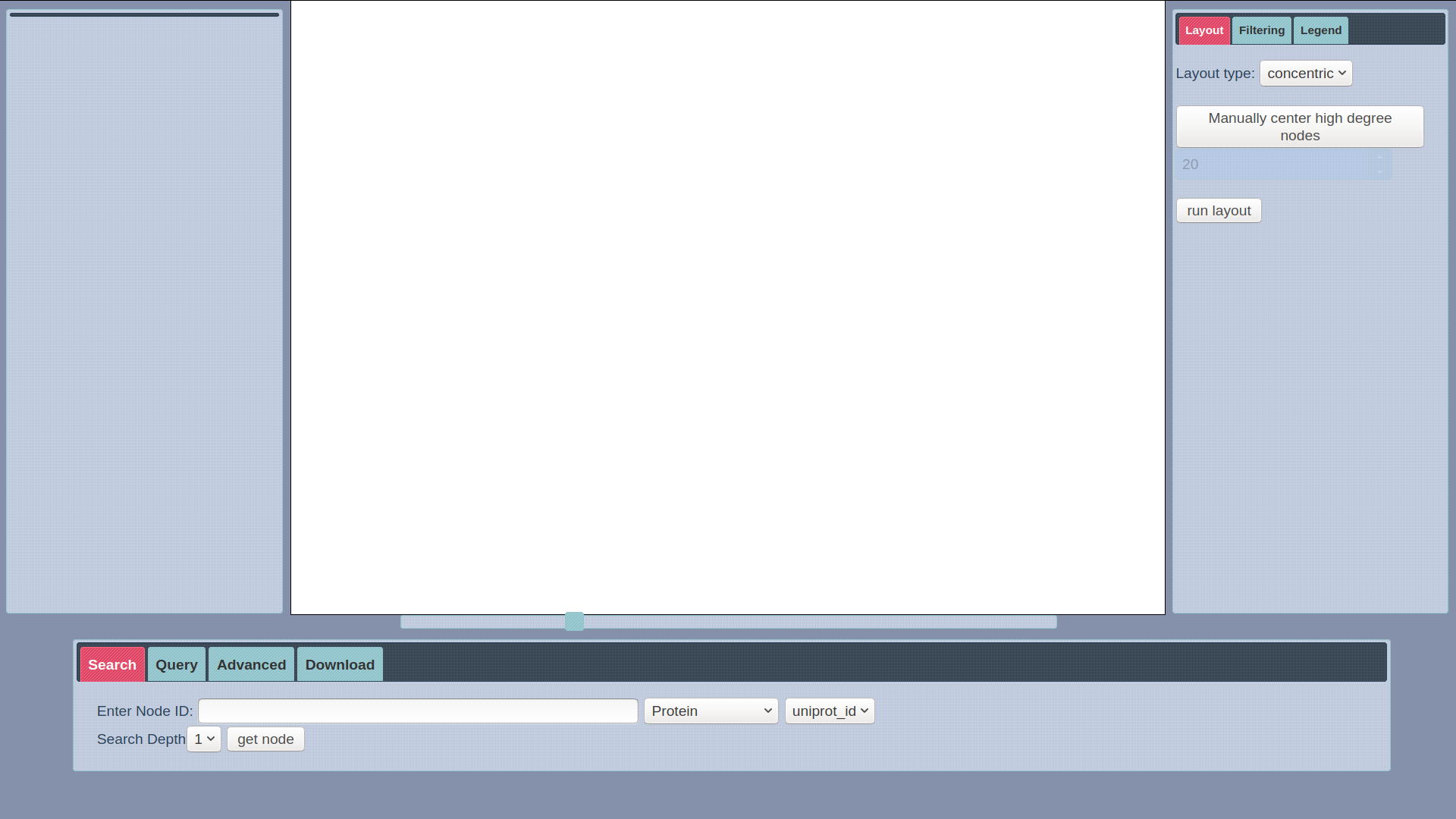
SmartBioSearch Interface
SmartBioSearch is a web application designed to provide a user-friendly querying, visualization and filtering interface for SmartBioGraph database. The application has a basic search functionality which automatically generates a cyper query to retrieve a node and its immediate neighbors, provided with the node label and search field. It has a visualization module based on Cytoscape with a custom concentric layout and other standard layout options. The interface can also support custom user queries written in CQL, provided that the resulting data from query is a graph. Click here to start your search.
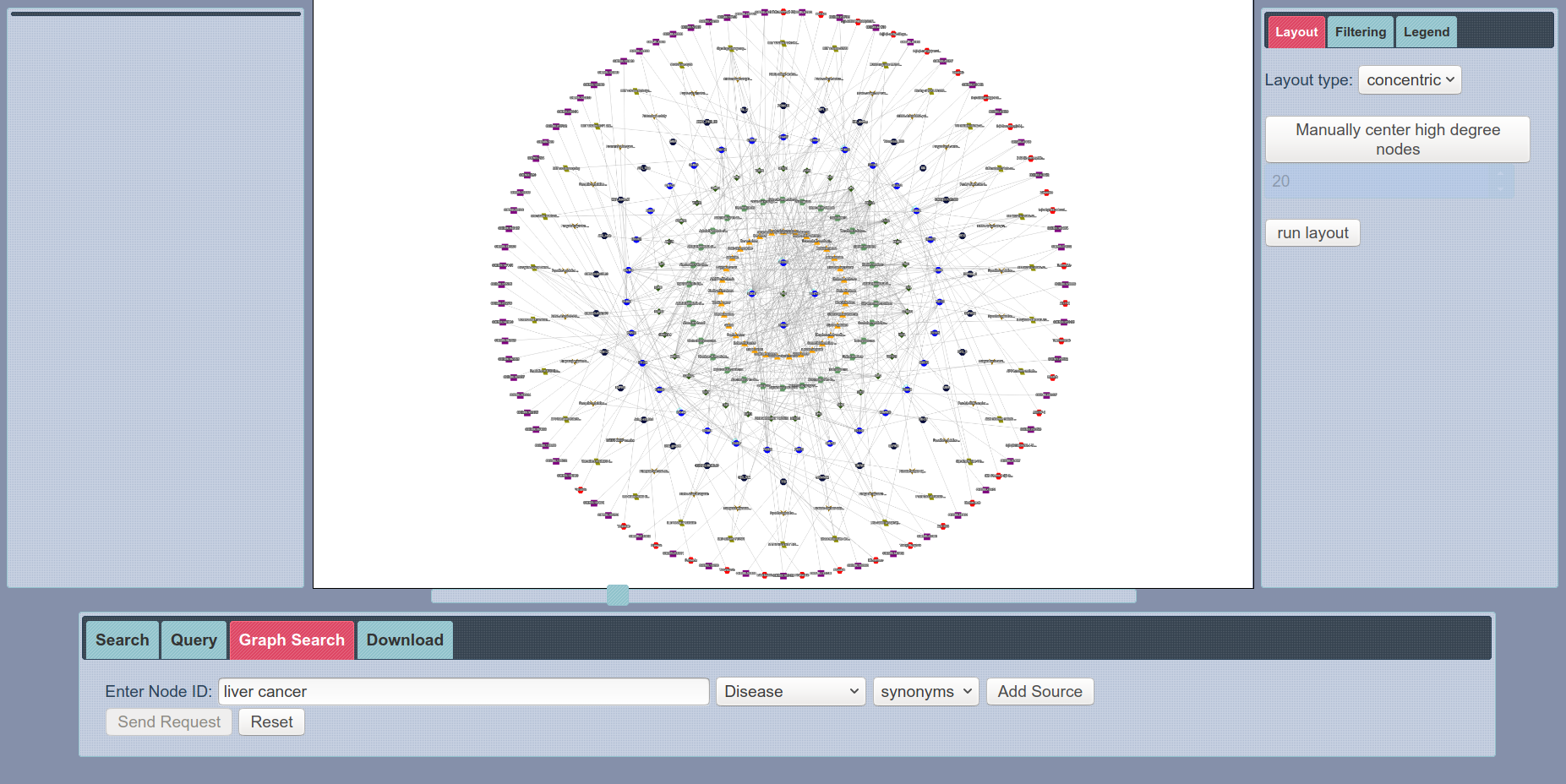
SmartBioSearch Interface Graph Search
Perhaps, the most exciting feature of SmartBioSearch interface is Graph Search, where users can search for a customized network topology with a easy-to-use network creator tool. Then, enrichment analysis is performed on the resulting graph from the query and nodes with less statistical significance are filtered out. This functionality enables users to obtain a statistically valuable biological network very easily, which would otherwise require a very detailed process. Check out our demo videos to see how this functionality can be used.
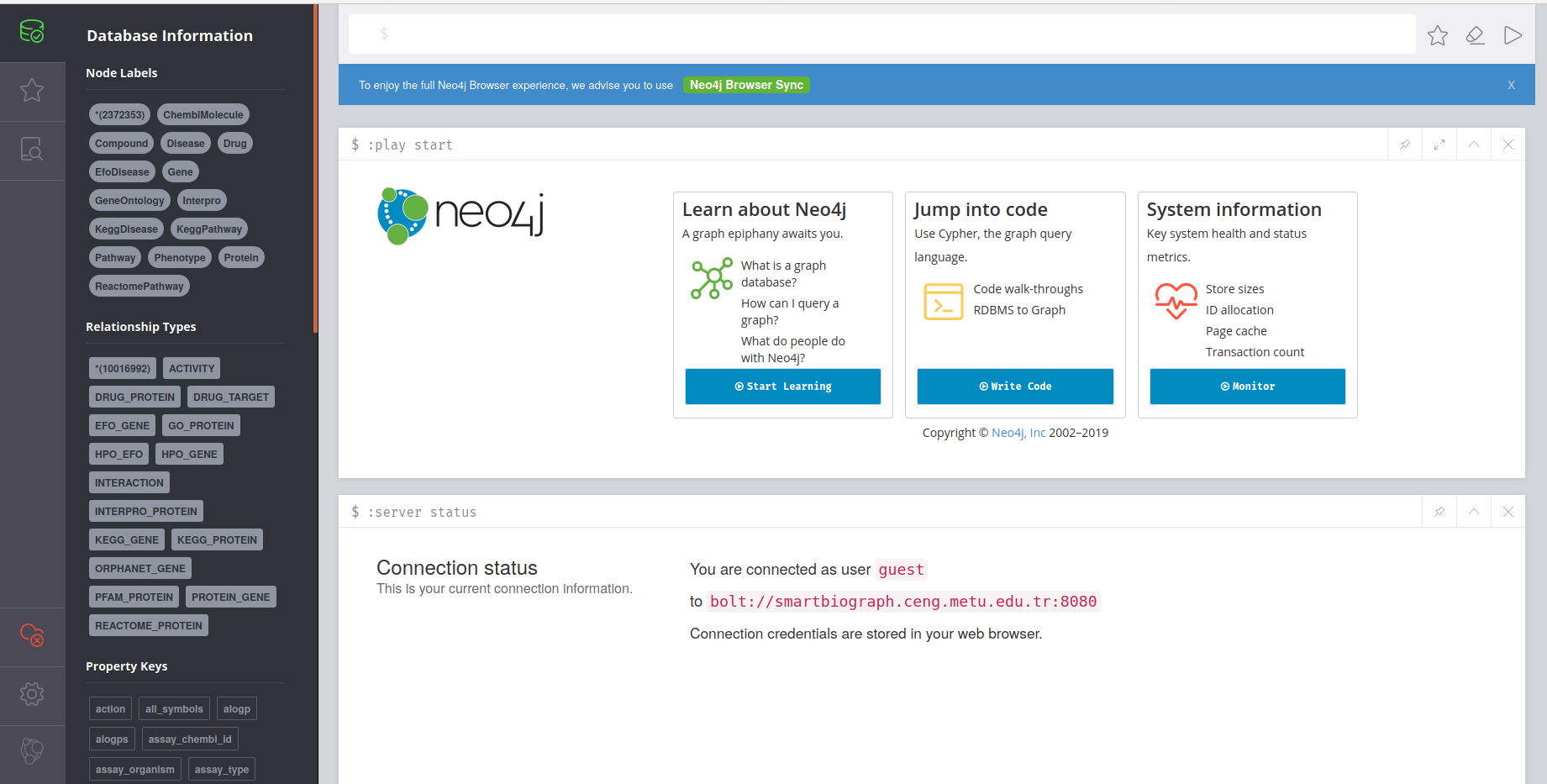
Neo4j Browser
Neo4J browser is a third party software that provides direct access to SmartBioGraph database. It is a great tool for visualization in small scale and for advanced custom user queries written in CQL. Neo4J Browser also has its own API to support software development on top of SmartBioGraph database.








